First ever nanoscale Hoberman structure built out of DNA origami
 Hoberman spheres are an example of “deployable structures,” which can change their size while maintaining their geometry. They can be simple mechanisms – like an umbrella or a scissor lift – or can contain complex geometry, such as the retractable roof of a stadium, or an unfolding satellite. But they’ve never been built at the nanoscale.
Hoberman spheres are an example of “deployable structures,” which can change their size while maintaining their geometry. They can be simple mechanisms – like an umbrella or a scissor lift – or can contain complex geometry, such as the retractable roof of a stadium, or an unfolding satellite. But they’ve never been built at the nanoscale.
“I had bought one of these Hoberman spheres for my kids,” said Jong Hyun Choi, associate professor of mechanical engineering. “I looked at it and thought, ‘I wonder if we can construct this with DNA?’”
Scientists use a technique called “DNA origami” to build nanoscale structures using synthetic DNA strands. Rather than using complex human DNA, they use a specific genome of bacteria known as M13mp18, whose DNA can be easily manipulated in a laboratory, and have its four basic building blocks – adenine (A), cytosine (C), guanine (G), and thymine (T) – programmed to connect in unique ways.
“We call it ‘constructing,’ but actually the DNA are assembling on their own,” said Choi. “The only thing this DNA knows how to do is connect A to T, and connect C to G. So if we give them a sequence of instructions that tells certain A’s to connect to certain T’s, and certain C’s to connect to certain G’s, the DNA molecules self-assemble into the shapes we want.”
Choi uses a standard CAD (computer-aided design) program to design the physical shapes. The software then spits out a sequence of A, C, G, and T characters, which Choi sends to an external company that sequences short strands complementary to M13mp18 bacterial DNA. After heating the mixture sample, Choi observes the resulting nanostructures using atomic force microscopy, because the shapes are about 1,000 times smaller than a human hair.
“The field of DNA origami is fascinating, but right now it’s all driven by chemistry,” said Choi. “I’m a mechanical engineer. I want to use DNA origami as a manufacturing system to build well-defined nanostructures, and observe their mechanical properties.”
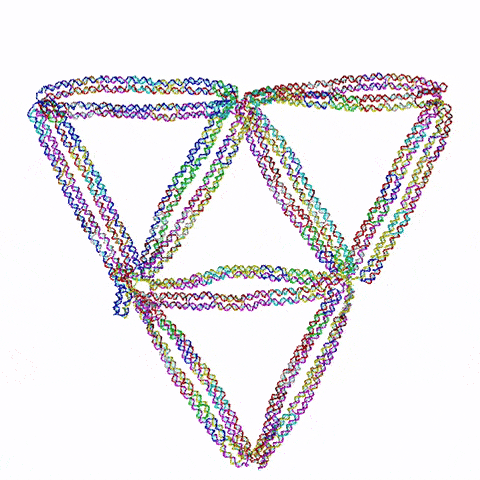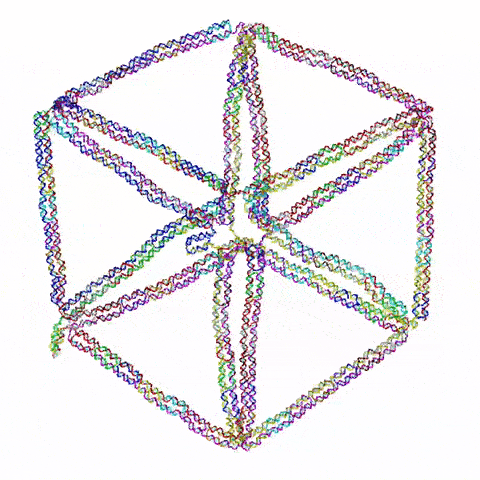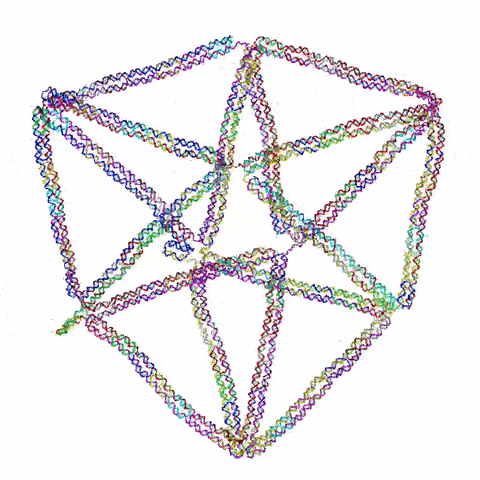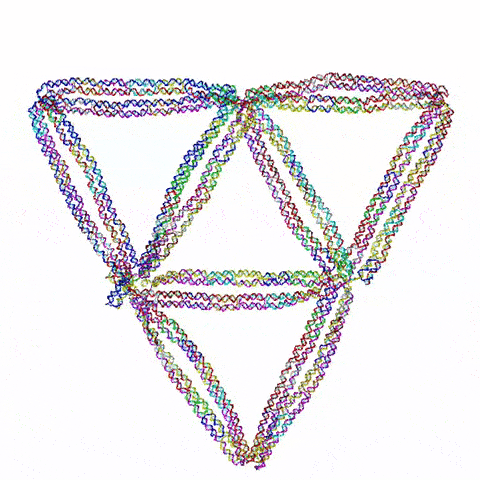
So the question becomes: what do you build? Choi’s team started with triangles, the simplest 2D shape possible. They then moved on to squares and simple polygons. Then they began connecting the shapes together. Soon they were experimenting with more complex topology, including architectured auxetic structures: fields of repeated shapes that move and fold in, thus showing unique mechanical properties distinct and different from regular materials.
One they proved that nanoscale deployable structures were possible, Choi’s team decided to build a Hoberman ring, a simplified 2D version of the Hoberman sphere. “They actually make a full-scale toy, called a Hoberman Flight Ring, which you can unfold and then throw like a Frisbee,” said Choi. “We were able to build a simple version of this structure with DNA origami, at less than 100 nanometers across – and show both states of its deployment.”
Their work has been published in the journals Small and Angewandte Chemie.
Choi admits that their research in this field is just beginning. For example, they have yet to observe the transformation of the Hoberman ring in process. “When you hold a Hoberman ring in your hands, you can push and pull on it to change its shape,” he said. “But at nanoscale, applying that mechanical force in real time is almost impossible.”
To simulate the open and closed states of the ring, they applied a tiny DNA strand to act as a “jack,” to pull on the edges and pry open the shape. In the microscopy, they were able to observe shapes with a short jack that were pried open, and shapes with a long jack that were still closed. “It’s tough to show the dynamics of what happens between those two states in real time,” said Choi. “We have the computer simulations, but we’re still working on how we can demonstrate that experimentally. Are there other ways to actuate these movements?”
They also have ambitions to build three dimensional structures. “3D is obviously more challenging,” said Choi, “and first we need to prove that it’s even possible with this DNA origami. But once we understand the mechanics, then we can construct design principles that allow us to build complex shapes and dynamic behaviors.”
Eventually, Choi sees big potential for using DNA origami as a manufacturing process. The most obvious applications are in biomedicine. “DNA, by its very nature, is biocompatible,” said Choi. “So these structures could be sent into parts of the body that traditional medicine can’t reach. They could be programmed to travel into the smallest blood vessels, and expand their shape to relieve stress and lower blood pressure. They could also act as sensors, to automatically actuate some process in plants or animals. That’s really our grand challenge.”
Writer: Jared Pike, jaredpike@purdue.edu, 765-496-0374
Source: Jong Hyun Choi, jchoi@purdue.edu, 765-496-3562
ABSTRACT
Topological Assembly of a Deployable Hoberman Flight Ring from DNA
Ruixin Li, Haorong Chen, and Jong Hyun Choi
https://doi.org/10.1002/smll.202007069
Deployable geometries are finite auxetic structures that preserve their overall shapes during expansion and contraction. The topological behaviors emerge from intricately arranged elements and their connections. Despite the considerable utility of such configurations in nature and in engineering, deployable nanostructures have never been demonstrated. Here a deployable flight ring, a simplified planar structure of Hoberman sphere is shown, using DNA origami. The DNA flight ring consists of topologically assembled six triangles in two layers that can slide against each other, thereby switching between two distinct (open and closed) states. The origami topology is a trefoil knot, and its auxetic reconfiguration results in negative Poisson’s ratios. This work shows the feasibility of deployable nanostructures, providing a versatile platform for topological studies and opening new opportunities for bioengineering.
Auxetic Two Dimensional Nanostructures from DNA
Ruixin Li, Haorong Chen, Jong Hyun Choi
https://doi.org/10.1002/anie.202014729
Architectured materials exhibit negative Poisson's ratios and enhanced mechanical properties compared with regular materials. Their auxetic behaviors emerge from periodic cellular structures regardless of the materials used. The majority of such metamaterials are constructed by topdown approaches and macroscopic with unit cells of microns or larger. There are also molecular auxetics including natural crystals which are not designable. There is a gap from few nanometers to microns, which may be filled by biomolecular self-assembly. Herein, we demonstrate two dimensional auxetic nanostructures using DNA origami. Structural reconfigurations are performed by two step DNA reactions and complemented by mechanical deformation studies using molecular dynamics simulations. We find that the auxetic behaviors are mostly defined by geometrical designs, yet the properties of the materials also play an important role. From elasticity theory, we introduce design principles for auxetic DNA metamaterials.
